Random walk
Qian Feng
2018-02-08
Last updated: 2018-02-08
Code version: fa85a0b
Random walk in 1,2,3 dimension examples, pretty interesting!
1D
##################################################
N = 10000
signal = vector(length = N)
signal[1] = 0
for (i in 2:N)
{
# random select one offset (from [-1,1]) to signal[i-1]
signal[i] = signal[i-1] + sample(c(-1,1),1)
}
plot( signal,type = 'l',col = 'yellow')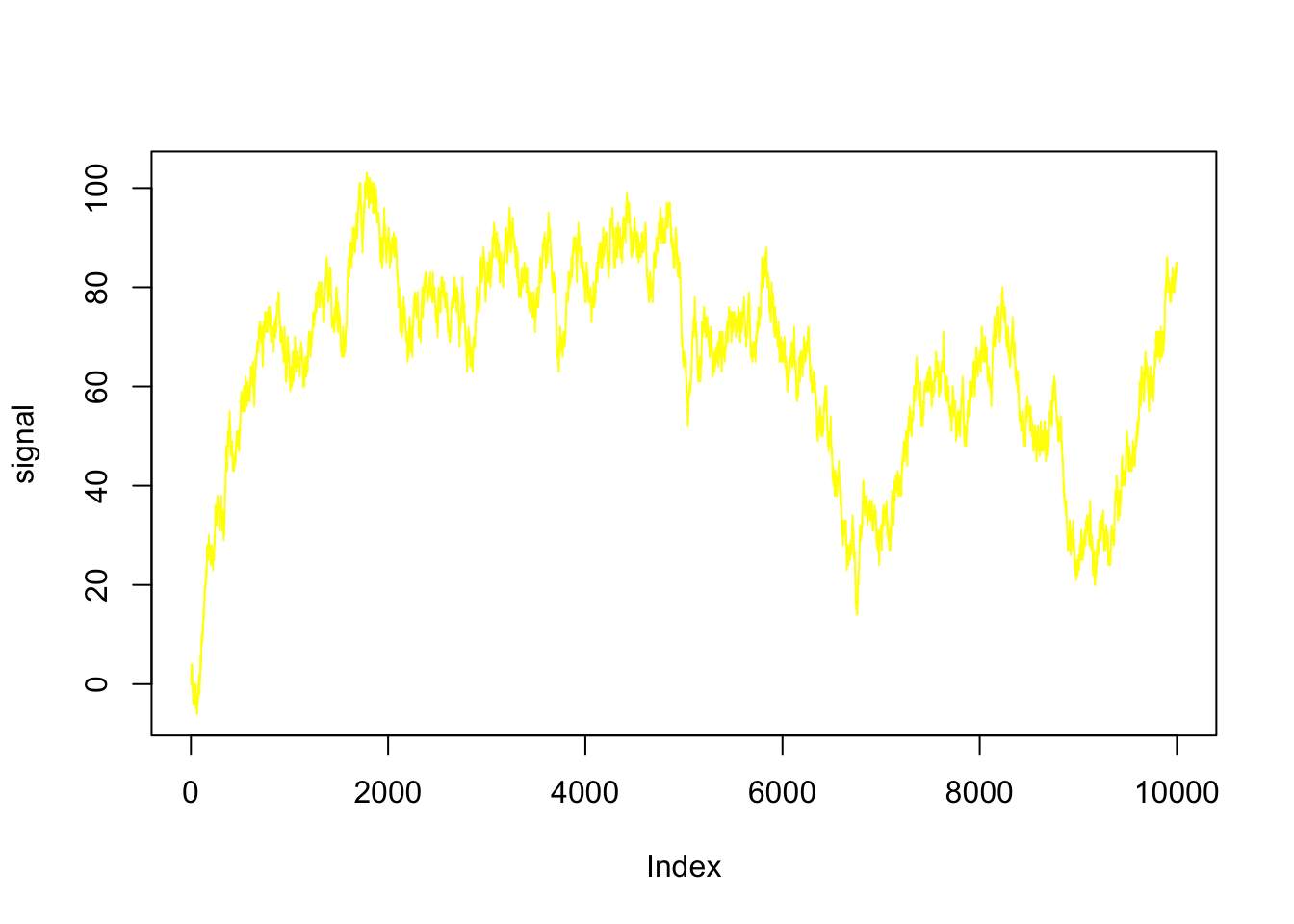
2D
##################################################
N = 100
x = vector(length = N)
y = vector(length = N)
x[1] = 0
y[1] = 0
for (i in 2:N)
{
x[i] = x[i-1] + rnorm(1)
y[i] = y[i-1] + rnorm(1)
}
plot(x,y,type = 'l', col='red')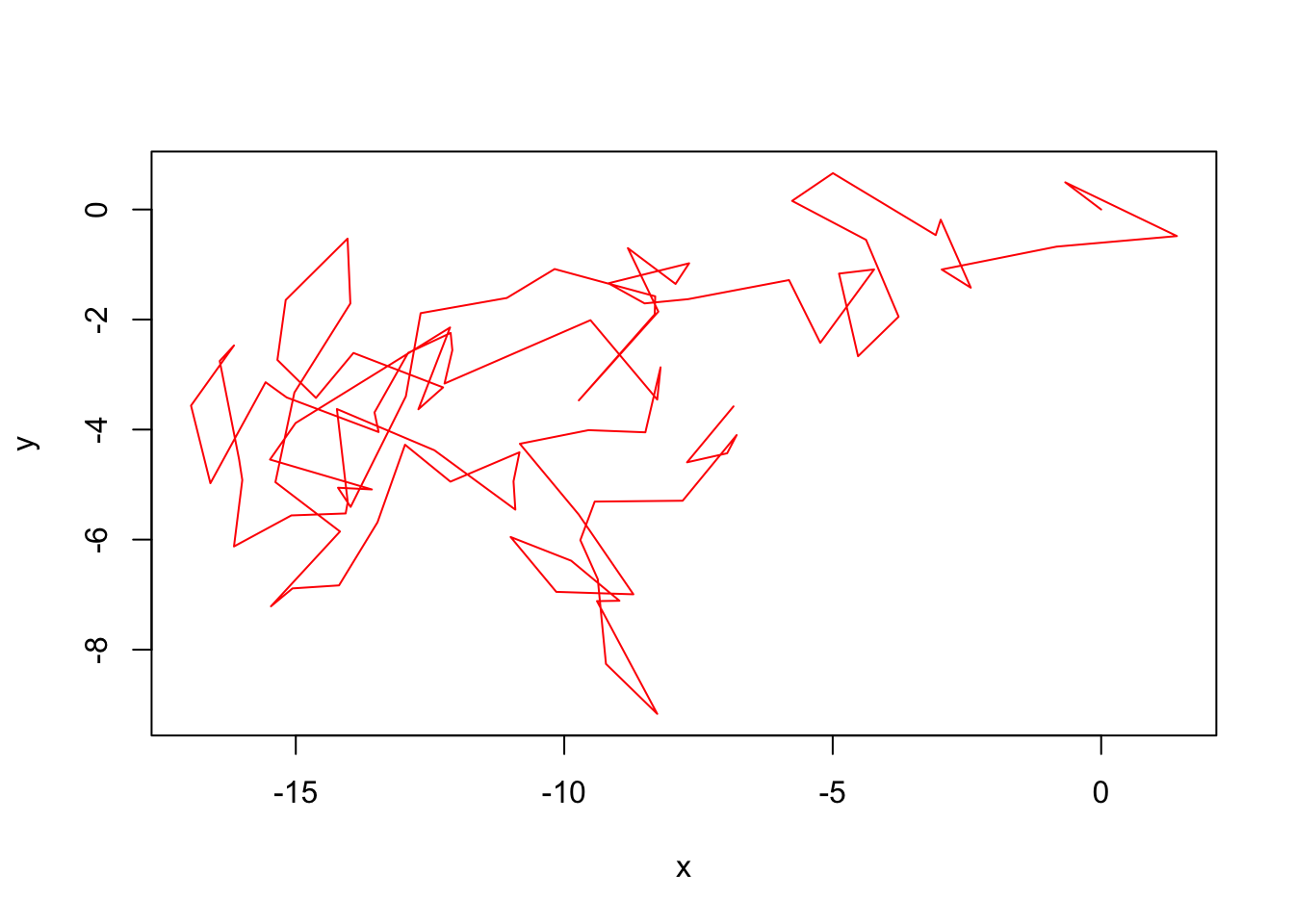
3D
##################################################
#source("http://bioconductor.org/biocLite.R")
#biocLite("scatterplot3d")
library("scatterplot3d")
N = 100
x = vector(length = N)
y = vector(length = N)
z = vector(length = N)
x[1] = 0
y[1] = 0
z[1] = 0
for (i in 2:N)
{
x[i] = x[i-1] + rnorm(1)
y[i] = y[i-1] + rnorm(1)
z[i] = z[i-1] + rnorm(1)
}
scatterplot3d(x, y, z,color="red", col.axis="blue", type="h", col.grid="lightblue",main="scatterplot3d - 1", pch=20)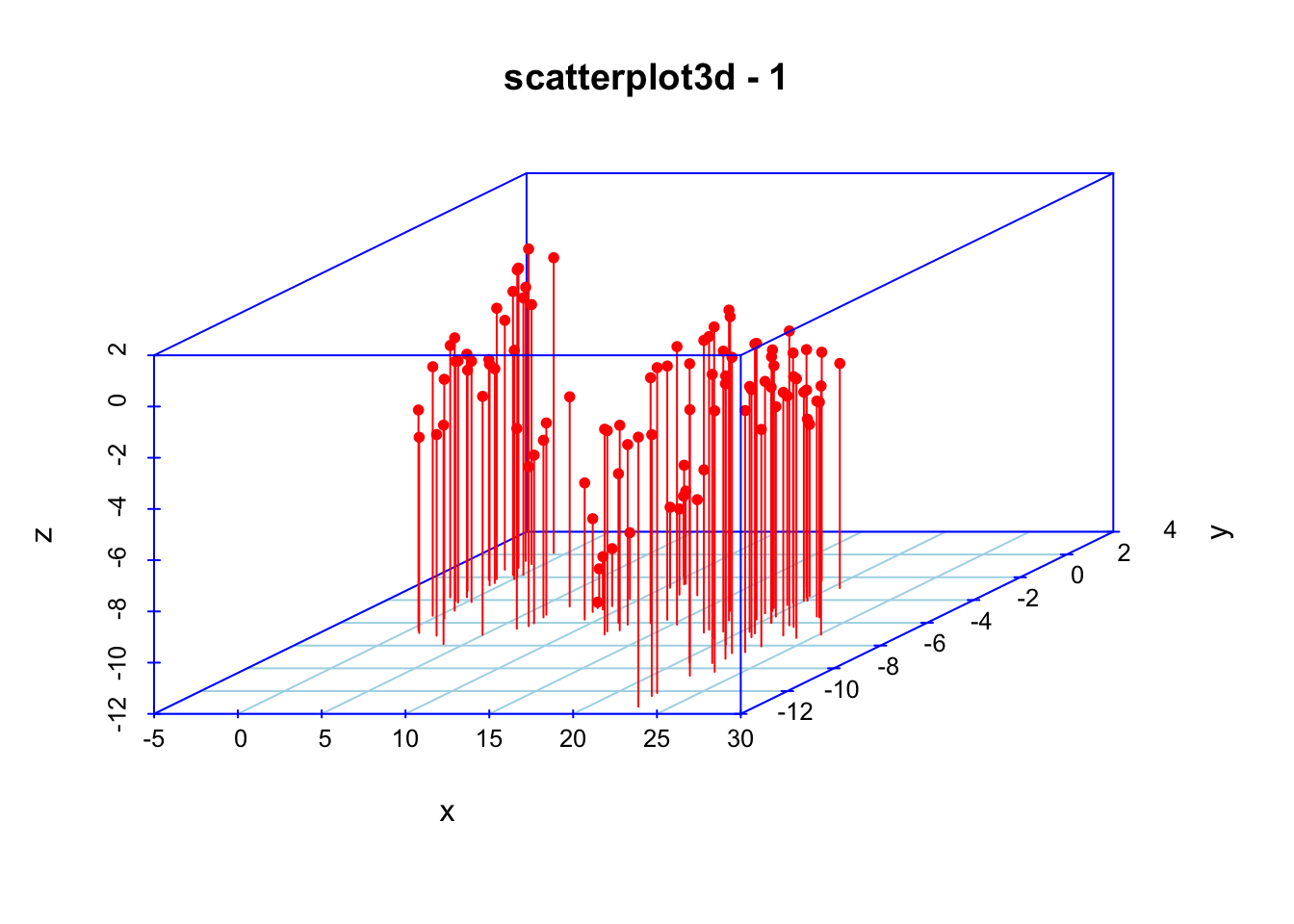
scatterplot3d(x, y, z,color="red", col.axis="blue", type="l", col.grid="lightblue",main="scatterplot3d - 1", pch=20)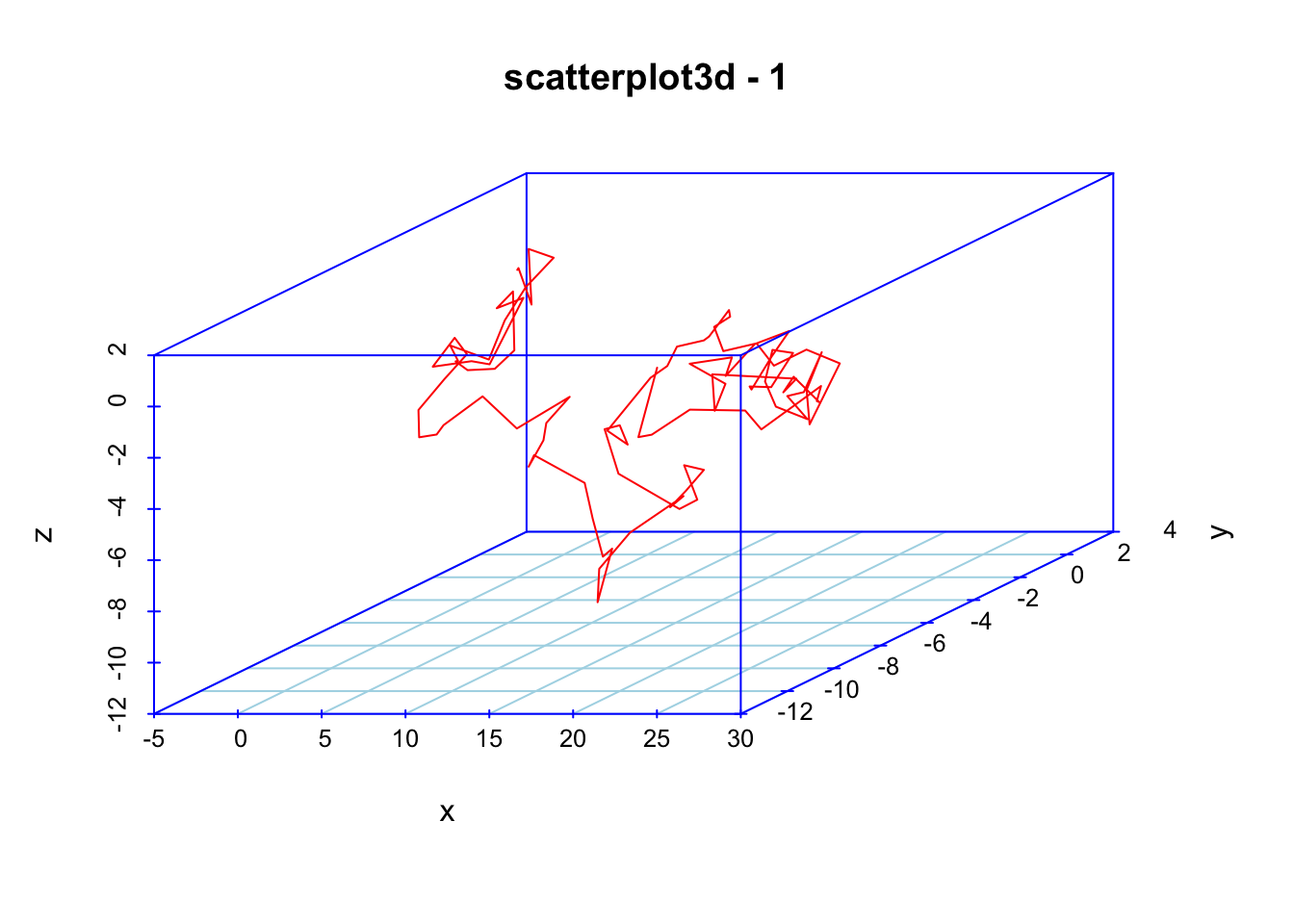
Session information
sessionInfo()R version 3.4.2 (2017-09-28)
Platform: x86_64-apple-darwin15.6.0 (64-bit)
Running under: macOS Sierra 10.12.1
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/3.4/Resources/lib/libRblas.0.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/3.4/Resources/lib/libRlapack.dylib
locale:
[1] zh_CN.UTF-8/zh_CN.UTF-8/zh_CN.UTF-8/C/zh_CN.UTF-8/zh_CN.UTF-8
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] scatterplot3d_0.3-40
loaded via a namespace (and not attached):
[1] compiler_3.4.2 backports_1.1.2 magrittr_1.5 rprojroot_1.3-2
[5] tools_3.4.2 htmltools_0.3.6 yaml_2.1.16 Rcpp_0.12.14
[9] stringi_1.1.6 rmarkdown_1.8 knitr_1.18 git2r_0.21.0
[13] stringr_1.2.0 digest_0.6.14 evaluate_0.10.1This R Markdown site was created with workflowr