Combinatorial methods for phylogenetic networks
Qian Feng
2018-01-04
Last updated: 2018-01-05
Code version: 582bc0b
Phylogenetic trees are not suitable to describe evolutionary history when datasets involve significantly plenty of reticulate events, including horizontal gene transfer, hybridization, recombination, reassortment etc. Phylogenetic network provides an alternative. It is any graph used to represent evolutionary relationships between a set of taxa that label some of its nodes.
This paper is a literature review, introducing briefly fundamental concepts about phylogenetic network and summarizing separate algorithms correspond to each type of network. Figure 1 vividly show all different types of phylogenetic network introduced in this essay. 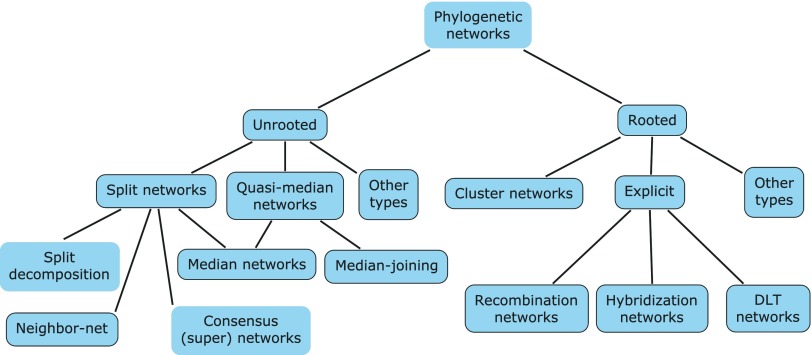 Figure 1 from original paper
Figure 1 from original paper
In theory, Phylogenetic network consists of two types: unrooted phylogenetic network and rooted phylogenetic network. The former one is much more widely used than the latter one in practice, because there are many problems needed to solve in rooted phylogenetic network. First, many algorithms are not designed as a tool in real studies though they have proof-of-concept implementations; second, algorithms have impractical running times. Therefore, developing suitable methods for rooted phylogenetic network is still a unforeseeable challenge.
From another point of view, phylogenetic networks are used in two ways, the first one is as a tool for visualizing incompatible clusters/taxa, we call it abstract, implicit or data-displayed networks; another one represents the evolutionary history including reticulate events, called explicit or evolutionary networks. In some sense, most unrooted phylogenetic networks are abstact, however, rooted phylogenetic networks could be either abstract or explicit.
\(\bigstar\) Pay attention to the difference among these words: Hybridization, Recombination, Mutation, Crossover, Duplication.I could not distinguish them actually.
Answers given by Yao-ban:Hybridization is about the species level. Recombination is about gene level. Crossover is double recombination. Mutation is change of specific gene. Duplication could repeat, like could make the gene longer, likewise, gene loss.
This R Markdown site was created with workflowr